Chair: Prof. Dr. Christian Freund, Freie Universität Berlin
Vice chair: Prof. Dr. Walter Nickel, Heidelberg University Biochemistry Center (BZH)
First funding period: July 2016 - June 2020
Second funding period: July 2020 - June 2024
Third funding period: July 2024 - June 2028
Summary:
A hallmark of cellular physiology is the coordination of signal transmission in space and time. Biological signals are generated by molecular switches cells employ to exert spatio-temporal control over a wide range of cellular processes. Prominent examples are protein secretion, endocytosis, receptor downstream signaling and circadian gene expression, among others. Typical examples for molecular switches include post-translational modifications produced by protein and lipid kinases, GTPases as well as calcium and redox switches. In this way, cells turn on and off signals that are required to coordinate cellular processes including responses to extracellular signals in space and time. On the one hand, this enables cells to dynamically organize molecular components of signaling processes in nanodomains and to localize a given downstream process. On the other hand, molecular switches can be used by cells in a way that enables them to operate processes at proper time scales. Intriguingly, a relatively small set of distinct
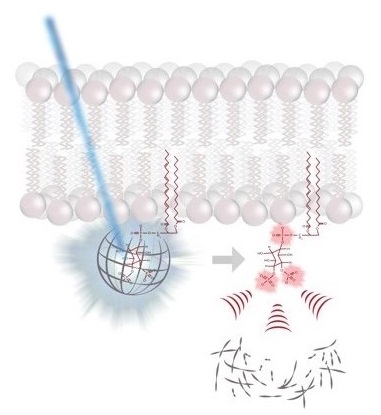 molecular switches is capable of controlling a large diversity of cellular processes with widely differing kinetics from milliseconds in neurotransmission to hours in circadian gene expression. While the molecular mechanisms by which molecular switches are turned on and off are known in great detail in many cases, the molecular mechanisms of how they coordinate the corresponding downstream processes in space and time are poorly understood. The overarching goal of our research consortium is to functionally analyze how a distinct set of molecular switches controls the spatio-temporal regulation of a diverse range of biological processes. Intriguingly, in many cases, the molecular events that are downstream of the activation of a molecular switch trigger a cellular switch such as cell fate decisions between survival and apoptosis. To investigate the molecular mechanisms that link molecular switches to cellular switches in living cells, we use advanced chemical biology tools such as photoactivatable membrane lipids. In addition, experimental setups are employed to synchronize protein transport through optogenetic control. Another example are approaches that allow for controlling protein activity by acute and reversible spatial sequestration of proteins of interest. These strategies are complemented by advanced biochemical reconstitution experiments, structural analyses as well as theoretical approaches such as kinetic modeling or molecular dynamics simulations. In this way, the dissection of the individual steps of a given process as determined by the assembly, activation and time-dependent localization of its key molecular components will continue to be a major goal of the TRR186 consortium. With this advanced set of interdisciplinary methodologies, our long-term goal is to obtain a comprehensive understanding of how signals generated by activated molecular switches translate into the precise spatio-temporal coordination of cellular processes such as protein secretion, receptor signaling, endocytosis and gene expression as well as other central activities that characterize living cells.
molecular switches is capable of controlling a large diversity of cellular processes with widely differing kinetics from milliseconds in neurotransmission to hours in circadian gene expression. While the molecular mechanisms by which molecular switches are turned on and off are known in great detail in many cases, the molecular mechanisms of how they coordinate the corresponding downstream processes in space and time are poorly understood. The overarching goal of our research consortium is to functionally analyze how a distinct set of molecular switches controls the spatio-temporal regulation of a diverse range of biological processes. Intriguingly, in many cases, the molecular events that are downstream of the activation of a molecular switch trigger a cellular switch such as cell fate decisions between survival and apoptosis. To investigate the molecular mechanisms that link molecular switches to cellular switches in living cells, we use advanced chemical biology tools such as photoactivatable membrane lipids. In addition, experimental setups are employed to synchronize protein transport through optogenetic control. Another example are approaches that allow for controlling protein activity by acute and reversible spatial sequestration of proteins of interest. These strategies are complemented by advanced biochemical reconstitution experiments, structural analyses as well as theoretical approaches such as kinetic modeling or molecular dynamics simulations. In this way, the dissection of the individual steps of a given process as determined by the assembly, activation and time-dependent localization of its key molecular components will continue to be a major goal of the TRR186 consortium. With this advanced set of interdisciplinary methodologies, our long-term goal is to obtain a comprehensive understanding of how signals generated by activated molecular switches translate into the precise spatio-temporal coordination of cellular processes such as protein secretion, receptor signaling, endocytosis and gene expression as well as other central activities that characterize living cells.
Please see the press release by the Heidelberg University's Communication and Marketing Press Office